Introduction to Small Molecule Binding
What is small molecule binding affinity?
Small molecule binding affinity describes the strength of the interaction between a small molecule and a target, often a protein.
Why determine the binding affintiy?
A strong binding affinity increases the probability that the clinical effect of the drug candidate is due to its interaction with the target, rather than with other structures in the organism.
Determining the binding affinity of a drug candidate helps you to:
- Understand the target of the drug candidate
- Make informed decisions that increase the probability of success in later phases of the drug discovery process.
It is important to determine the binding affinity early in the drug discovery process.
info
The small molecule binding affinity should be determined before moving forward with in-vivo experiments. This will help ensure that only candidates with strong binding affinity to their target protein are selected for further development.
Predicting binding affinity
Machine Learning models can be used to predict the binding affinity of a small molecule and protein.
At LabDAO we are focused on curating and maintaining the most effective tools to accelerate the drug discovery process. You can browse the available tools and run a tutorial to get started.
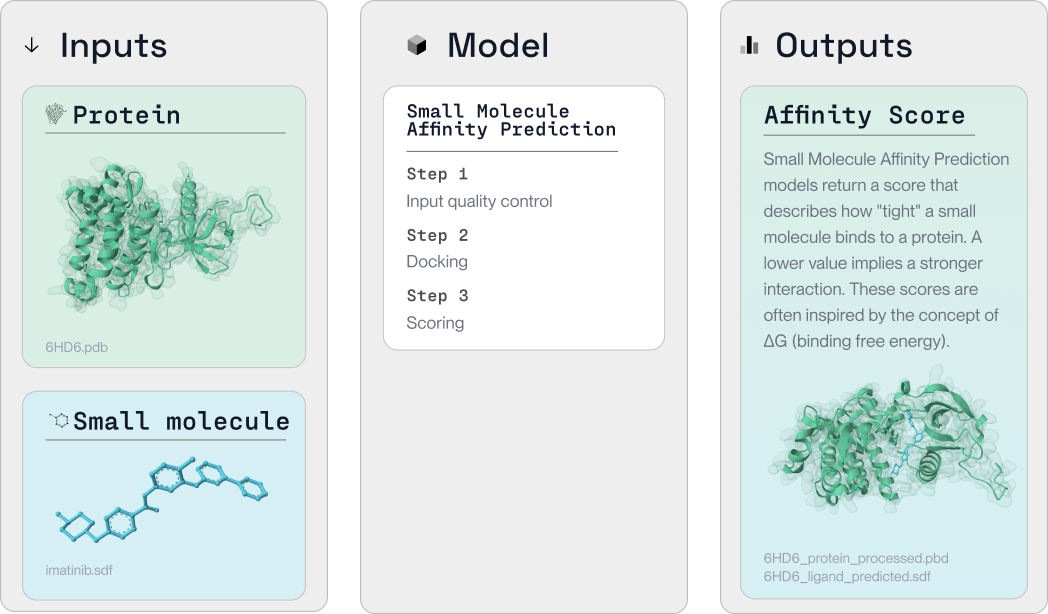
When predicting binding affinity using LabDAO's computational tools, the process is as follows:
Input: Small molecule(s) and protein(s)
Model:
- Step 1: Quality control checks
- Step 2: Dock the small molecule and protein using molecular docking tools e.g. Diffdock
- Step 3: Score the predicted interaction, to determine how tightly the small molecule and protein are expected to bind to each other
Outputs: The model will return the score, as well as 2 separate files showing the small molecule and the protein - these can be used to explore the interaction visually
Describing binding affinity
The binding affinity of a small molecule and a protein can be described with three related metrics:
- ΔG (binding free energy)
- Kd (dissociation constant)
- IC50 (half-maximal inhibitory concentration).
Generally, ΔG is considered the most generalisable metric to describe binding affinity. IC50 is easy to measure, but it is a highly context dependent metric.
For each of these three metrics, a lower value implies a "tighter" interaction between the small molecule and target protein.
How to interpret the binding affintiy?
Binding affinity data can be interpreted by comparing the ΔG, Kd, or IC50 values to a set of reference values for the target. References can be taken from previous experiments with known binders or by determining the binding affinity for a series of candidates and comparing them with eachother.
For all three binding affinity metrics, a smaller score indicates a stronger binding affinity. A strong interaction is indicated by:
- A negative ΔG value
- Kd or IC50 close to 0
caution
Predicting the binding affinity with machine learning models should be interpreted with additional care. It is important to consider the reported accuracy of these models, as well as their underlying training data, when reviewing results.
Measuring binding affinity in the lab
There are several laboratory methods used to measure the binding affinity of a protein and small molecule. Popular, accurate and label-free methods include Surface-Plasmon-Resonance (SPR) and Bio-layer Interferometry (BLI). The cost of an average SPR experiment can range from several thousand dollars to tens of thousands of dollars, depending on the complexity of the experiment and the equipment used.
TODO: get in touch with to access more than 40 laboratories offering Surface-Plasmon-Resonance